| LEF1 |
|---|
 |
|
| Dynodwyr |
|---|
| Cyfenwau | LEF1, LEF-1, TCF10, TCF1ALPHA, TCF7L3, lymphoid enhancer binding factor 1 |
|---|
| Dynodwyr allanol | HomoloGene: 7813 GeneCards: LEF1 |
|---|
|
| Ontoleg y genyn |
|---|
| Gweithrediad moleciwlaidd | • GO:0001131, GO:0001151, GO:0001130, GO:0001204 DNA-binding transcription factor activity
• GO:0001077, GO:0001212, GO:0001213, GO:0001211, GO:0001205 DNA-binding transcription activator activity, RNA polymerase II-specific
• histone deacetylase binding
• armadillo repeat domain binding
• estrogen receptor binding
• RNA polymerase II transcription regulatory region sequence-specific DNA binding
• transcription factor binding
• estrogen receptor activity
• gamma-catenin binding
• GO:0031493 histone binding
• C2H2 zinc finger domain binding
• chromatin binding
• cysteine-type endopeptidase inhibitor activity involved in apoptotic process
• GO:0001948, GO:0016582 protein binding
• DNA binding, bending
• DNA binding
• sequence-specific DNA binding
• transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding
• beta-catenin binding
• GO:0000980 RNA polymerase II cis-regulatory region sequence-specific DNA binding
• GO:0001200, GO:0001133, GO:0001201 DNA-binding transcription factor activity, RNA polymerase II-specific
|
|---|
| Cydrannau o'r gell | • cytoplasm
• cnewyllyn cell
• transcription regulator complex
• beta-catenin-TCF complex
• nucleoplasm
• protein-DNA complex
|
|---|
| Prosesau biolegol | • somitogenesis
• tongue development
• paraxial mesoderm formation
• GO:0044324, GO:0003256, GO:1901213, GO:0046019, GO:0046020, GO:1900094, GO:0061216, GO:0060994, GO:1902064, GO:0003258, GO:0072212 regulation of transcription by RNA polymerase II
• negative regulation of DNA binding
• T cell receptor V(D)J recombination
• transcription by RNA polymerase II
• cellular response to cytokine stimulus
• mammary gland development
• negative regulation of interleukin-13 production
• odontogenesis of dentin-containing tooth
• apoptotic process involved in blood vessel morphogenesis
• cell chemotaxis
• negative regulation of canonical Wnt signaling pathway
• face morphogenesis
• neutrophil differentiation
• negative regulation of interleukin-5 production
• GO:0009373 regulation of transcription, DNA-templated
• dentate gyrus development
• apoptotic process involved in morphogenesis
• transcription, DNA-templated
• GO:0060469, GO:0009371 positive regulation of transcription, DNA-templated
• positive regulation of cell growth
• regulation of Wnt signaling pathway
• embryonic limb morphogenesis
• hypothalamus development
• branching involved in blood vessel morphogenesis
• trachea gland development
• regulation of striated muscle tissue development
• positive regulation of cell-cell adhesion
• forebrain neuroblast division
• eye pigmentation
• negative regulation of apoptotic process
• sprouting angiogenesis
• GO:1901227 negative regulation of transcription by RNA polymerase II
• positive regulation of epithelial to mesenchymal transition
• BMP signaling pathway
• positive regulation of cell cycle process
• osteoblast differentiation
• histone H3 acetylation
• formation of radial glial scaffolds
• positive regulation of granulocyte differentiation
• alpha-beta T cell differentiation
• canonical Wnt signaling pathway
• GO:0045996 negative regulation of transcription, DNA-templated
• sensory perception of taste
• cellular response to interleukin-4
• negative regulation of cysteine-type endopeptidase activity involved in apoptotic process
• cell development
• negative regulation of cell-cell adhesion
• negative regulation of interleukin-4 production
• negative regulation of striated muscle tissue development
• positive regulation of cell migration
• Wnt signaling pathway
• T-helper 1 cell differentiation
• positive regulation of cell proliferation in bone marrow
• anatomical structure regression
• chorio-allantoic fusion
• negative regulation of apoptotic process in bone marrow cell
• B cell proliferation
• GO:1901313 positive regulation of gene expression
• vasculature development
• forebrain radial glial cell differentiation
• neural crest cell migration
• positive regulation of cell population proliferation
• forebrain neuron differentiation
• histone H4 acetylation
• hippocampus development
• GO:0003257, GO:0010735, GO:1901228, GO:1900622, GO:1904488 positive regulation of transcription by RNA polymerase II
• Wnt signaling pathway, calcium modulating pathway
• steroid hormone mediated signaling pathway
• odontoblast differentiation
• beta-catenin-TCF complex assembly
• epithelial to mesenchymal transition
• regulation of cell-cell adhesion
• secondary palate development
|
|---|
| Sources:Amigo / QuickGO |
|
| Orthologau |
|---|
| Species | Bod dynol | Llygoden |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | | |
|---|
| RefSeq (protein) | | |
|---|
| Lleoliad (UCSC) | n/a | n/a |
|---|
| PubMed search | [1] | n/a |
|---|
| Wicidata |
|
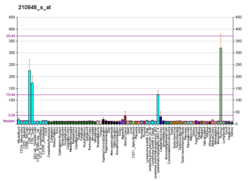
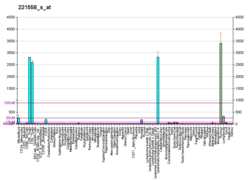
Protein sy'n cael ei godio yn y corff dynol gan y genyn LEF1 yw LEF1 a elwir hefyd yn Lymphoid enhancer-binding factor 1, isoform CRA_b a Lymphoid enhancer binding factor 1 (Saesneg). Segment o DNA yw'r genyn, sy'n amgodio ffwythiant arbennig. Mae'r genyn yma wedi ei leoli ar yr edefyn ôl o gromosom dynol 4, band 4q25.[2]
Yn aml mae gan enynnau lawer o gyfystyron. Mae hyn oherwydd eu bod yn aml yn cael eu darganfod gan nifer o bobl mewn cyd-destunau gwahanol heb wybod mai'r un genynnau oeddyn nhw. Hefyd mae gan wahanol gymunedau gwyddonol safonau gwahanol ar gyfer enwi genynnau. Dyma restr o gyfystyron ar gyfer y genyn LEF1.
- LEF-1
- TCF10
- TCF7L3
- TCF1ALPHA
- "Methylprednisolone suppresses the Wnt signaling pathway in chronic lymphocytic leukemia cell line MEC-1 regulated by LEF-1 expression. ". Int J Clin Exp Pathol. 2015. PMID 26339357.
- "Overexpression of lymphoid enhancer-binding factor-1 (LEF1) is a novel favorable prognostic factor in childhood acute lymphoblastic leukemia. ". Int J Lab Hematol. 2015. PMID 25955539.
- "Diagnostic Utility of Lymphoid Enhancer Binding Factor 1 Immunohistochemistry in Small B-Cell Lymphomas. ". Am J Clin Pathol. 2017. PMID 28395058.
- "Expression of LEF1 in mantle cell lymphoma. ". Ann Diagn Pathol. 2017. PMID 28038713.
- "High LEF1 expression predicts adverse prognosis in chronic lymphocytic leukemia and may be targeted by ethacrynic acid.". Oncotarget. 2016. PMID 26950276.