保存配列

進化生物学において、保存配列 (conserved sequence) とは、種間にまたがって、もしくはゲノム中に存在する、類似した、もしくは同一の核酸(DNAとRNA)・タンパク質配列のことである。種間で保存されている配列をオルソログ(オーソログ)配列、ゲノム中で保存されている複数の配列をパラログ配列と呼ぶ。保存性は、その配列が自然選択の過程で維持されてきたことを意味する。
高度に保存された配列は、系統樹をはるかにさかのぼっても比較的変わらないままであり、それゆえ地質時代的なタイムスケールにおいても同様である。高度に保存された配列の例は、生命のすべてのドメインに存在するリボソームのRNA要素、真核生物に広く存在するホメオボックス、そして細菌のtmRNAなどである。配列保存性の研究は、ゲノミクス、プロテオミクス、進化生物学、系統学、バイオインフォマティクス、そして数学の研究分野と重なる。
配列保存性の研究の歴史
[編集]DNAの遺伝における役割の発見と、1949年のフレデリック・サンガーによる動物種間のインスリンの差異についての報告[2]は、初期の分子生物学者たちを分子レベルの観点からの系統学の研究へと促した[3][4] 。 1960年代、DNAハイブリダイゼーションやタンパク質の交差反応性 (cross-reactivity)に基づいた研究によって、ヘモグロビン[5]やシトクロムc[6] のような既知のオルソログタンパク質間の類似性が測定された。1965年、Émile Zuckerkandl とライナス・ポーリングは分子時計の概念を導入し[7] 、定常的な突然変異率を2つの生物が分枝してからの時間の推定のために用いることができると提唱した。初期の系統発生は化石記録とよく一致している一方で、いくつかの遺伝子については異なる速度で進化しているように思われ、そこから分子進化の理論のさらなる発展がもたらされた[3][4] 。1966年のMargaret Dayhoff によるフェレドキシンの配列比較は、自然選択は生命に必須のタンパク質の配列を保存し最適化するように作用する、と示した[8]。
配列保存のメカニズム
[編集]いくつもの世代にわたって、ある進化系統のゲノム中の核酸配列はランダムな変異や欠失によって、時間経過と共に徐々に変化していく[9][10]。また、配列は染色体の再編成 (chromosomal rearrangement) によって組み替えられたり欠失したりすることもある。保存配列は、このような変化への圧力にもかかわらずゲノム中に存在し続ける配列であり、突然変異率がバックグラウンドレベルよりも低い配列である[11]。
核酸配列の保存は、コーディング領域であってもノンコーディング領域であっても起こり得る。高度に保存されたDNA配列は何らかの機能的な価値を持つと考えられているが、多くの高度に保存されたノンコーディングなDNA配列の役割はほとんどわかっていない。配列が保存される程度は、選択圧や、突然変異に対する頑強性 (robustness)、集団サイズや遺伝的浮動の影響を受ける。
コーディング配列
[編集]DNA上にコードされているアミノ酸は3塩基が1組で決定されており、塩基はATGCの4種類であるのに対して、DNA上にコードされているアミノ酸は20種類である。したがって、理論上DNA上にコード可能なアミノ酸の種類よりも少なく、別な塩基配列であっても同じアミノ酸をコードしている場合もある。コーディグ領域に変異が発生しても、タンパク質のアミノ酸配列に影響しない、同義的な変異をサイレント変異と言う。サイレント変異が起こっても、作られるタンパク質は同じなので、このような変異が起きたとしても影響が少ないため、保存性は低くなりがちである。
アミノ酸配列は、タンパク質またはタンパク質ドメインの構造や機能を維持するよう保存される。保存されたタンパク質はアミノ酸の置換が少ないか、もしくは類似した生化学的性質を持つアミノ酸への置換が起きやすい傾向を持つ。配列の中でも、フォールディング、構造安定性、もしくは結合部位を形成するアミノ酸は、タンパク質の機能に変化をもたらし得るため、より高度に保存される。
タンパク質をコードする遺伝子の核酸配列は他の選択圧によっても保存される。生物種によるコドンの使用頻度の偏り (codon usage bias) は、配列中の同義置換のタイプを制限する。mRNAで二次構造を形成する核酸配列は、翻訳に悪影響を与えるものもあるため、不利な選択を受ける。一方でそのmRNAが機能的なノンコーディングRNAとしても機能する場合には保存される[12][13]。
ノンコーディング配列
[編集]ノンコーディング配列は、リボソームや転写因子の結合部位や認識部位となるなど、遺伝子発現の調節に重要であり、ゲノム中で保存されうる。例えば、保存された遺伝子やオペロンでは、そのプロモーター領域もまた保存される。タンパク質と同様に、ノンコーディングRNA (ncRNA) においても構造と機能に重要な核酸は保存される。しかしながら、タンパク質をコードする配列と比較して、ncRNAにおける配列保存性は一般的に乏しく、その代わり、構造や機能に寄与する塩基対の位置がしばしば保存されている[14][15]。例えば、インターロイキン22の遺伝子の付近にも、そのような場所が見られる[16]。
保存配列の同定
[編集]保存配列は、一般的にシーケンスアラインメントに基づいたバイオインフォマティクスによるアプローチによって同定される。ハイスループットなDNAシークエンシングやタンパク質質量分析の手法の進歩によって、2000年代初期以降、比較可能なタンパク質配列やゲノム情報の量は格段に増加した。
相同性検索(ホモロジー・サーチ)
[編集]保存配列は、BLASTやHMMER、Infernal[17]などのツールを用いた相同性検索によって同定することができる。相同性検索ツールは、個々の核酸またはタンパク質の配列をインプットとして用いてもよいし、既知の関連する配列の多重アラインメントから生成された統計モデルを用いてもよい。プロファイルHMMや構造情報を組み込んだRNA共分散モデル[18]のような統計モデルは、より関連性の低い配列を検索する場合に有用である。入力された配列は、関連個体または他の種の配列のデータベースに対してアラインメントされる。その結果は、一致するアミノ酸の数や、アラインメントによって生じたギャップや欠失の数に基づいてスコアリングがなされる。許容される保存的な置換は、PAMやBLOSUMのような置換行列に基づいて同定される。高スコアのアラインメントは、相同配列からなるものと推定される。
多重配列アラインメント
[編集]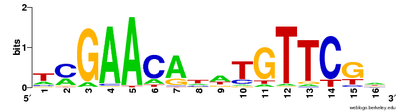
多重配列アラインメントは保存配列を可視化するためにも用いられる。Clustalフォーマットにはアラインメント中の保存残基の位置に注釈をつけるプレーンテキストのキーがあり、保存された残基の位置がアスタリスク(*) で、保存的な変異の位置がコロン(:) で、準保存的な変異の位置がピリオド(.)で、非保存的な変異の位置が空欄( )でそれぞれ示される[20](ページ上部の図参照)。 シーケンスロゴも、アラインメント中の各位置における残基の割合を高さによって表すことで、保存配列を表現する。
ゲノムアラインメント
[編集]ホールゲノムアラインメント (whole genome alignment, WGA) もまた、種間で高度に保存された領域を同定するために用いられる。現段階では、組み換えや反復配列、そして真核生物のゲノムの大きなサイズを扱う計算の複雑さのために、WGAのツールの正確さやスケーラビリティには限界がある[21]。しかしながら、30以上の近縁の細菌についてのWGAが実現可能なものとなっている[22][23]。
スコアリングシステム
[編集]他のアプローチでは、統計的な検定に基づいた保存性の測定が用いられ、予測されるバックグラウンドの(中立な)突然変異率とは異なる変異率を示す領域の同定が試みられている。
GERP (Genomic Evolutionary Rate Profiling) フレームワークは、種間の遺伝的配列の保存性を評価する。このアプローチでは、多重配列アラインメントからある生物種のセットにおける中立的な突然変異率を推定し、その予測値よりも変異が少ない配列領域を同定する。そして、これらの領域は予測されるバックグラウンドの突然変異率と観測された実際の突然変異率との差に基づいてスコアリングがなされる。高いGERPスコアは、高度に保存された配列であることを意味する[24][25]。
PhyloP や PhyloHHM のようなアプローチでは、置換率の確率分布を比較する統計系統学 (statistical phylogenetics) 的な手法が組み込まれており、変異の保存と加速の両方を検出することができる。はじめに、系統樹に基づいて、多重配列アラインメント中のあるカラムで起こると予測される置換の数のバックグラウンドの確率分布が生成される。目的の種間の進化的関係は、任意の置換の有意性を計算するために用いられる。すなわち、きわめて近縁の種間の変異は遠縁の種間の変異よりも起こりにくく、それゆえより有意である。保存性を検出するため、多重配列アラインメントの一部に対して確率分布が計算され、尤度比検定またはスコア検定のような手法を用いてバックグラウンド分布との比較が行われる。そして、2つの分布を比較して生成されたP値をもとに、保存領域が同定される[26][27][28]。
極端な保存性
[編集]超保存エレメント
[編集]超保存エレメント(ultra-conserved element, UCE) は、複数の分類群にわたって高度に類似した、もしくは同一の配列である。UCEは最初に脊椎動物の間に見つかり[29]、続いて次第に幅広く異なる分類群の間で同定された[30]。UCEの起源や機能はほとんどわかっていないが[31]、有羊膜類[32]、昆虫[33]、そして動物と植物[34]、といった地質学的ともいえるタイムスケールにおける差異を調べる際に利用されてきた。
普遍的に保存された遺伝子
[編集]最も高度に保存された遺伝子は、すべての生物に見つかる遺伝子である。それらは、主に転写や翻訳に必要とされるノンコーディングRNAやタンパク質であり、全生物の共通祖先 (LUCA) から保存されていると考えられている[35]。
普遍的に保存されていることが判明している遺伝子もしくは遺伝子ファミリーには、GTP結合型翻訳伸長因子、メチオニルアミノペプチダーゼ2、セリンヒドロキシメチルトランスフェラーゼ、ABC輸送体が含まれる[36]。RNAポリメラーゼやヘリカーゼといった転写装置の構成要素や、rRNA、tRNA、リボソームタンパク質といった翻訳装置の構成要素もまた、普遍的に保存されている[37]。
応用
[編集]系統発生学と分類学
[編集]保存された配列のセットは、しばしば系統樹を作成するために用いられる。なぜなら、類似した配列を持つ生物種は近縁関係にあると考えられるからである[38]。どのような配列を選択すべきかは、その分類学的研究の視野によって異なる。例えば、最も高度に保存されている16S RNAや他のリボソームの配列は、系統樹の根元に近い部分の関係性を再構築したり、メタゲノム解析において細菌の門を同定する場合などに有用である[39][40]。ある系統群で保存されている一方でいくつかの変異があるような配列、例えばハウスキーピング遺伝子などは、生物種間の関係性を調べるために用いられる[41][42][43]。ITS (internal transcribed spacer) 領域は、保存されたrRNA遺伝子の間のスペーサーとして必要であるが急速な進化を経ており、菌類の分類や急速に進化する細菌の菌株の分類によく用いられる[44][45][46][47]。
医学研究
[編集]高度に保存された配列はしばしば重要な生物学的機能を有しており、遺伝疾患の原因を特定する際のスタート地点として有用である。多くの先天性代謝異常症やライゾソーム病は個々の保存された遺伝子が変化した結果であり、疾患の症状の根本的要因となる酵素の欠失または欠損が引き起こされている。遺伝疾患は、マウスやショウジョウバエといった実験動物とヒトの間で保存されている配列を特定し、その遺伝子をノックアウトしたときの影響を調べることで予測することができる[48][49][50]。またゲノムワイド関連解析も、疾患や health outcome と関連した保存配列の変化を同定するために用いられる[51][52]。
機能のアノテーション
[編集]保存配列の同定は、遺伝子のような機能的配列を発見したり予測したりするためにも用いられる[53]。タンパク質ドメインのような既知の機能を持つ保存配列が、配列から機能を予測する際には用いられる。Pfam や Conserved Domain Database といった保存されたタンパク質ドメインのデータベースが、タンパク質をコードすると予測された遺伝子の中に機能的ドメインをアノテーションするために用いられる[54]。
出典
[編集]- ^ “Clustal FAQ #Symbols”. Clustal. 8 December 2014閲覧。
- ^ Sanger, F. (24 September 1949). “Species Differences in Insulins”. Nature 164 (4169): 529–529. doi:10.1038/164529a0.
- ^ a b Marmur, J; Falkow, S; Mandel, M (October 1963). “New Approaches to Bacterial Taxonomy”. Annual Review of Microbiology 17 (1): 329–372. doi:10.1146/annurev.mi.17.100163.001553.
- ^ a b Pace, N. R.; Sapp, J.; Goldenfeld, N. (17 January 2012). “Phylogeny and beyond: Scientific, historical, and conceptual significance of the first tree of life”. Proceedings of the National Academy of Sciences 109 (4): 1011–1018. doi:10.1073/pnas.1109716109. PMC 3268332.
- ^ Zuckerlandl, Emile; Pauling, Linus B. (1962). “Molecular disease, evolution, and genetic heterogeneity”. Horizons in Biochemistry: 189–225.
- ^ Margoliash, E (Oct 1963). “PRIMARY STRUCTURE AND EVOLUTION OF CYTOCHROME C”. Proc Natl Acad Sci U S A 50 (4): 672–679. doi:10.1073/pnas.50.4.672. PMC 221244.
- ^ Zuckerkandl, E; Pauling, LB (1965). “Evolutionary Divergence and Convergence in Proteins”. Evolving Genes and Proteins: 96–166. doi:10.1016/B978-1-4832-2734-4.50017-6.
- ^ Eck, R. V.; Dayhoff, M. O. (15 April 1966). “Evolution of the Structure of Ferredoxin Based on Living Relics of Primitive Amino Acid Sequences”. Science 152 (3720): 363–366. doi:10.1126/science.152.3720.363.
- ^ Kimura, M (17 February 1968). “Evolutionary Rate at the Molecular Level”. Nature 217 (5129): 624–626. doi:10.1038/217624a0.
- ^ King, J. L.; Jukes, T. H. (16 May 1969). “Non-Darwinian Evolution”. Science 164 (3881): 788–798. doi:10.1126/science.164.3881.788.
- ^ Kimura, M; Ohta, T (1974). “On Some Principles Governing Molecular Evolution”. Proc Natl Acad Sci USA 71 (7): 2848–2852. doi:10.1073/pnas.71.7.2848. PMC 388569. PMID 4527913.
- ^ Chamary, JV; Hurst, Laurence D (2005). “Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals”. Genome Biology 6 (9): R75. doi:10.1186/gb-2005-6-9-r75.
- ^ Wadler, C. S.; Vanderpool, C. K. (27 November 2007). “A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide”. Proceedings of the National Academy of Sciences 104 (51): 20454–20459. doi:10.1073/pnas.0708102104. PMC 2154452.
- ^ Johnsson, Per; Lipovich, Leonard; Grandér, Dan; Morris, Kevin V. (March 2014). “Evolutionary conservation of long non-coding RNAs; sequence, structure, function”. Biochimica et Biophysica Acta (BBA) - General Subjects 1840 (3): 1063–1071. doi:10.1016/j.bbagen.2013.10.035. PMC 3909678.
- ^ Freyhult, E. K.; Bollback, J. P.; Gardner, P. P. (6 December 2006). “Exploring genomic dark matter: A critical assessment of the performance of homology search methods on noncoding RNA”. Genome Research 17 (1): 117–125. doi:10.1101/gr.5890907. PMC 1716261.
- ^ 機能食品成分によるT細胞分化のエピジェネティクス制御機構の解明
- ^ Nawrocki, E. P.; Eddy, S. R. (4 September 2013). “Infernal 1.1: 100-fold faster RNA homology searches”. Bioinformatics 29 (22): 2933–2935. doi:10.1093/bioinformatics/btt509. PMC 3810854.
- ^ Eddy, SR; Durbin, R (11 June 1994). “RNA sequence analysis using covariance models.”. Nucleic Acids Research 22 (11): 2079–88. doi:10.1093/nar/22.11.2079. PMC 308124. PMID 8029015.
- ^ “Weblogo”. UC Berkeley. 30 December 2017閲覧。
- ^ “Clustal FAQ #Symbols”. Clustal. 8 December 2014閲覧。
- ^ Earl, Dent; Nguyen, Ngan; Hickey, Glenn; Harris, Robert S.; Fitzgerald, Stephen; Beal, Kathryn; Seledtsov, Igor; Molodtsov, Vladimir et al. (December 2014). “Alignathon: a competitive assessment of whole-genome alignment methods”. Genome Research 24 (12): 2077–2089. doi:10.1101/gr.174920.114.
- ^ Rouli, L.; Merhej, V.; Fournier, P.-E.; Raoult, D. (September 2015). “The bacterial pangenome as a new tool for analysing pathogenic bacteria”. New Microbes and New Infections 7: 72–85. doi:10.1016/j.nmni.2015.06.005.
- ^ Méric, Guillaume; Yahara, Koji; Mageiros, Leonardos; Pascoe, Ben; Maiden, Martin C. J.; Jolley, Keith A.; Sheppard, Samuel K.; Bereswill, Stefan (27 March 2014). “A Reference Pan-Genome Approach to Comparative Bacterial Genomics: Identification of Novel Epidemiological Markers in Pathogenic Campylobacter”. PLoS ONE 9 (3): e92798. doi:10.1371/journal.pone.0092798.
- ^ Cooper, G. M. (17 June 2005). “Distribution and intensity of constraint in mammalian genomic sequence”. Genome Research 15 (7): 901–913. doi:10.1101/gr.3577405.
- ^ “Sidow Lab - GERP”. mendel.stanford.edu. 2018年10月29日閲覧。
- ^ Pollard, K. S.; Hubisz, M. J.; Rosenbloom, K. R.; Siepel, A. (26 October 2009). “Detection of nonneutral substitution rates on mammalian phylogenies”. Genome Research 20 (1): 110–121. doi:10.1101/gr.097857.109.
- ^ “PHAST: Home”. compgen.cshl.edu. 2018年10月29日閲覧。
- ^ Fan, Xiaodan; Zhu, Jun; Schadt, Eric E; Liu, Jun S (2007). “Statistical power of phylo-HMM for evolutionarily conserved element detection”. BMC Bioinformatics 8 (1): 374. doi:10.1186/1471-2105-8-374.
- ^ Bejerano, G. (28 May 2004). “Ultraconserved Elements in the Human Genome”. Science 304 (5675): 1321–1325. doi:10.1126/science.1098119.
- ^ Siepel, A. (1 August 2005). “Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes”. Genome Research 15 (8): 1034–1050. doi:10.1101/gr.3715005.
- ^ Harmston, N.; Baresic, A.; Lenhard, B. (11 November 2013). “The mystery of extreme non-coding conservation”. Philosophical Transactions of the Royal Society B: Biological Sciences 368 (1632): 20130021–20130021. doi:10.1098/rstb.2013.0021.
- ^ Faircloth, B. C.; McCormack, J. E.; Crawford, N. G.; Harvey, M. G.; Brumfield, R. T.; Glenn, T. C. (9 January 2012). “Ultraconserved Elements Anchor Thousands of Genetic Markers Spanning Multiple Evolutionary Timescales”. Systematic Biology 61 (5): 717–726. doi:10.1093/sysbio/sys004.
- ^ Faircloth, Brant C.; Branstetter, Michael G.; White, Noor D.; Brady, Seán G. (May 2015). “Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among Hymenoptera”. Molecular Ecology Resources 15 (3): 489–501. doi:10.1111/1755-0998.12328.
- ^ Reneker, J.; Lyons, E.; Conant, G. C.; Pires, J. C.; Freeling, M.; Shyu, C.-R.; Korkin, D. (10 April 2012). “Long identical multispecies elements in plant and animal genomes”. Proceedings of the National Academy of Sciences 109 (19): E1183–E1191. doi:10.1073/pnas.1121356109.
- ^ Isenbarger, Thomas A.; Carr, Christopher E.; Johnson, Sarah Stewart; Finney, Michael; Church, George M.; Gilbert, Walter; Zuber, Maria T.; Ruvkun, Gary (14 October 2008). “The Most Conserved Genome Segments for Life Detection on Earth and Other Planets”. Origins of Life and Evolution of Biospheres 38 (6): 517–533. doi:10.1007/s11084-008-9148-z.
- ^ Harris, J. K. (12 February 2003). “The Genetic Core of the Universal Ancestor”. Genome Research 13 (3): 407–412. doi:10.1101/gr.652803.
- ^ Ban, Nenad; Beckmann, Roland; Cate, Jamie HD; Dinman, Jonathan D; Dragon, François; Ellis, Steven R; Lafontaine, Denis LJ; Lindahl, Lasse et al. (February 2014). “A new system for naming ribosomal proteins”. Current Opinion in Structural Biology 24: 165–169. doi:10.1016/j.sbi.2014.01.002.
- ^ Gadagkar, Sudhindra R.; Rosenberg, Michael S.; Kumar, Sudhir (15 January 2005). “Inferring species phylogenies from multiple genes: Concatenated sequence tree versus consensus gene tree”. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 304B (1): 64–74. doi:10.1002/jez.b.21026.
- ^ Ludwig, W; Schleifer, KH (October 1994). “Bacterial phylogeny based on 16S and 23S rRNA sequence analysis.”. FEMS Microbiology Reviews 15 (2–3): 155–73. doi:10.1111/j.1574-6976.1994.tb00132.x. PMID 7524576.
- ^ Hug, Laura A.; Baker, Brett J.; Anantharaman, Karthik; Brown, Christopher T.; Probst, Alexander J.; Castelle, Cindy J.; Butterfield, Cristina N.; Hernsdorf, Alex W. et al. (11 April 2016). “A new view of the tree of life”. Nature Microbiology 1 (5): 16048. doi:10.1038/nmicrobiol.2016.48.
- ^ Zhang, Liqing; Li, Wen-Hsiung (February 2004). “Mammalian Housekeeping Genes Evolve More Slowly than Tissue-Specific Genes”. Molecular Biology and Evolution 21 (2): 236–239. doi:10.1093/molbev/msh010.
- ^ Clermont, O.; Bonacorsi, S.; Bingen, E. (1 October 2000). “Rapid and Simple Determination of the Escherichia coli Phylogenetic Group”. Applied and Environmental Microbiology 66 (10): 4555–4558. doi:10.1128/AEM.66.10.4555-4558.2000.
- ^ Kullberg, Morgan; Nilsson, Maria A.; Arnason, Ulfur; Harley, Eric H.; Janke, Axel (August 2006). “Housekeeping Genes for Phylogenetic Analysis of Eutherian Relationships”. Molecular Biology and Evolution 23 (8): 1493–1503. doi:10.1093/molbev/msl027.
- ^ Schoch, C. L.; Seifert, K. A.; Huhndorf, S.; Robert, V.; Spouge, J. L.; Levesque, C. A.; Chen, W.; Bolchacova, E. et al. (27 March 2012). “Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi”. Proceedings of the National Academy of Sciences 109 (16): 6241–6246. doi:10.1073/pnas.1117018109.
- ^ Man, S. M.; Kaakoush, N. O.; Octavia, S.; Mitchell, H. (26 March 2010). “The Internal Transcribed Spacer Region, a New Tool for Use in Species Differentiation and Delineation of Systematic Relationships within the Campylobacter Genus”. Applied and Environmental Microbiology 76 (10): 3071–3081. doi:10.1128/AEM.02551-09.
- ^ Ranjard, L.; Poly, F.; Lata, J.-C.; Mougel, C.; Thioulouse, J.; Nazaret, S. (1 October 2001). “Characterization of Bacterial and Fungal Soil Communities by Automated Ribosomal Intergenic Spacer Analysis Fingerprints: Biological and Methodological Variability”. Applied and Environmental Microbiology 67 (10): 4479–4487. doi:10.1128/AEM.67.10.4479-4487.2001.
- ^ Bidet, Philippe; Barbut, Frédéric; Lalande, Valérie; Burghoffer, Béatrice; Petit, Jean-Claude (June 1999). “Development of a new PCR-ribotyping method for based on ribosomal RNA gene sequencing”. FEMS Microbiology Letters 175 (2): 261–266. doi:10.1111/j.1574-6968.1999.tb13629.x.
- ^ Ala, Ugo; Piro, Rosario Michael; Grassi, Elena; Damasco, Christian; Silengo, Lorenzo; Oti, Martin; Provero, Paolo; Di Cunto, Ferdinando et al. (28 March 2008). “Prediction of Human Disease Genes by Human-Mouse Conserved Coexpression Analysis”. PLoS Computational Biology 4 (3): e1000043. doi:10.1371/journal.pcbi.1000043.
- ^ Pandey, U. B.; Nichols, C. D. (17 March 2011). “Human Disease Models in Drosophila melanogaster and the Role of the Fly in Therapeutic Drug Discovery”. Pharmacological Reviews 63 (2): 411–436. doi:10.1124/pr.110.003293.
- ^ Huang, Hui; Winter, Eitan E; Wang, Huajun; Weinstock, Keith G; Xing, Heming; Goodstadt, Leo; Stenson, Peter D; Cooper, David N et al. (2004). Genome Biology 5 (7): R47. doi:10.1186/gb-2004-5-7-r47.
- ^ Ge, Dongliang; Fellay, Jacques; Thompson, Alexander J.; Simon, Jason S.; Shianna, Kevin V.; Urban, Thomas J.; Heinzen, Erin L.; Qiu, Ping et al. (16 August 2009). “Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance”. Nature 461 (7262): 399–401. doi:10.1038/nature08309.
- ^ Bertram, L. (2009). “Genome-wide association studies in Alzheimer's disease”. Human Molecular Genetics 18: R137–R145. doi:10.1093/hmg/ddp406.
- ^ Kellis, Manolis; Patterson, Nick; Endrizzi, Matthew; Birren, Bruce; Lander, Eric S. (15 May 2003). “Sequencing and comparison of yeast species to identify genes and regulatory elements”. Nature 423 (6937): 241–254. doi:10.1038/nature01644.
- ^ Marchler-Bauer, A.; Lu, S.; Anderson, J. B.; Chitsaz, F.; Derbyshire, M. K.; DeWeese-Scott, C.; Fong, J. H.; Geer, L. Y. et al. (24 November 2010). “CDD: a Conserved Domain Database for the functional annotation of proteins”. Nucleic Acids Research 39 (Database): D225–D229. doi:10.1093/nar/gkq1189.
関連項目
[編集]